Glossary of Terms Used in Combinatorial
Chemistry
[A-B]
[C] [D] [E-G]
[H-K] [L]
[M-N] [O-P] [Q] [R]
[S] [T] [U-Z]
D
Decode: Use of a surrogate analyte to define
the reaction path to which the solid support was exposed, and hence
imply the structure of a member of a combinatorial
library (see also encoding),
or the reaction sequence for its preparation 21.
Deconvolute: To render less complex;
Process of optimizing an activity of interest by fractionating (normally
by resynthesis, or by elaborating
a partial library)
a pool with some level of the desired
activity to give a set of smaller pools. Repeating this strategy leads
to single members with (ideally) a high level of activity and is termed
iterative deconvolution 22-24.
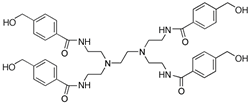
Dendrimer: A polymer having a
regular branched structure; If suitably functionalized (such as the
benzyl alcohol-substituted, four-branch structure shown below) may be
used as a soluble support, in
which case the desired, dendrimer-supported, material may be isolated
by size-exclusion chromatography. Dendrimers may also be attached
to a polymer and used as a solid support,
with significantly increased loading
over the initial resin 25-28.
Descriptor: Numerical representation
of a molecular property, including bulk properties (e.g. logP,
molecular weight), two-dimensional (2-D) features (atom connectivities),
or three-dimensional (3-D) features (molecular shape). A complete set
of descriptors comprises a fingerprint
16,29,30.
See also binning, clustering.
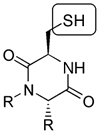
Directed Library: (also biased-
or focused library). Library
which uses a limited number of building
blocks chosen on the basis of preexisting information or hypothesis
which defines the type of functionalities deemed important to obtain
a particular activity. For example, every member of the diketopiperazine
library shown below contains the thiol pharmacophore
known to interact with metalloproteinase enzymes 31-33.
See also Unbiased Library.
Direct Divide: Strategy for the
assembly of a combinatorial
library related to the pool/split
process in which each portion of solid support is divided into the next
set of reaction vessels without an intervening pooling step. The resulting
library is (like a pool/split library) fully
combinatorial, with each particle bearing a single library member,
but has a reduced standard deviation between the quantity of each library
member 34.
Directed Sorting: Technique for
organizing a mixture of solid-supported samples by identifying each
particle ( for instance, on the basis of its shape, marking, or by reading
a radiofrequency code)
and transferring it to an appropriate position in an array. See also
sort and combine 35,36.
Diversity: The "unrelatedness"
of a set of, for example, building
blocks or members of a combinatorial
library, as measured by their properties such as atom connectivity,
physical properties, computational measurements, or bioactivity 17,37.
Diversity Reagent: One of a
set of reagents which introduces diversity
into the library products, as opposed to one which results in an identical
conversion for each member of the library. Similar to building
block but may be useful to distinguish from other (i.e. 'non-diversity')
reagents.
Divide, Couple, Recombine: see Pool/Split
Dynamic Library: Collection of
compounds in dynamic equilibrium. If the composition of the library
is altered, for instance by the presence of a receptor which selectively
binds certain library members,
then shifting of the equilibrium will lead to an increase in the amount
of those of components which bind to the target with relatively high
affinity 38,39.

